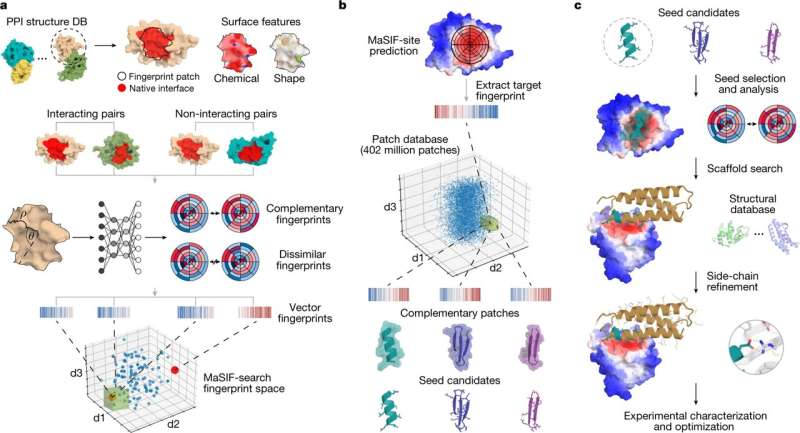
Solving the mystery of protein surface interactions with geometric fingerprints
Researchers from the Swiss Institute of Bioinformatics, Lausanne in Switzerland, used a geometric deep-learning tool that generates "fingerprints" of protein surfaces to describe geometric and chemical features critical to protein–protein interactions. In their paper, "De novo design of protein interactions with learned surface fingerprints," published in Nature, the team report that their hypothesized "fingerprints" led to the capture of essential aspects of molecular recognition and novel protein interactions. A Research Briefing that summarizes the team's results is published in the same journal issue.
Proteins are the physical machinery of biology, the cogs and gears, springs and valves that allow organic life to function. It is how cells do work, how drugs interact with biological systems and where most diseases interact with biological systems. With a commanding mastery of this machinery, science could cure most pathologies.
Machine learning, along with proteomics, genomic sequencing and molecular biology, have massively accelerated protein research over the past decade to the point where we can predict nearly all existing functional structures, engineer novel structures in any configuration and synthetically manufacture any protein imaginable with one tiny but significant caveat—the binding sites.
The way proteins interact relies on two specific physical "lock-and-key" interactions based on surface chemistry and structure. There is an outer rim site and a buried site beneath. The buried site does the work of the protein, but to access it, the initiating outer rim signal is required to open the structure to allow access.
As far as proteomics has come in the past decade, the affinity characteristics of proteins have remained elusive. This is partly because proteins are finicky and pH-dependent, with changeable rim surface chemistry and binding sites that are condition and site-specific. Read More…



